Storing data in DNA is a lot easier than getting it back out
Humanity is creating information at an unprecedented rate—some 16 zettabytes every year (a zettabyte is one billion terabytes). And this rate is increasing. Last year, the research group IDC calculated that we’ll be producing over 160 zettabytes every year by 2025.
All this data has to be stored, and as a result we need much denser memory than we have today. One intriguing solution is to exploit the molecular structure of DNA. Researchers have long known that DNA can be used for data storage—after all, it stores the blueprint for making individual humans and transmits it from one generation to the next.
What’s impressive for computer scientists is the density of the data that DNA stores: a single gram can hold roughly a zettabyte.
But nobody has come up with a realistic system for storing data in a DNA library and then retrieving it again when it is needed.
Today that changes thanks to the work of Federico Tavella at the University of Padua in Italy and colleagues, who have designed and tested just such a technique based on bacterial nanonetworks.
The principle is simple. Bacteria often carry genetic information in the form of tiny circular rings of double-stranded DNA called plasmids. These molecules are important because they often confer some advantage to the host cell, such as antibiotic resistance.
Crucially, bacteria can transfer plasmids from one cell to another in a process known as conjugation. This is one way that bacteria swap genetic information, and the process forms a fantastically complex nanonetwork in nature.
That’s the basis of the new technique. Tavella and co want to exploit this nanonetwork to transfer information that they have genetically engineered into the plasmids.
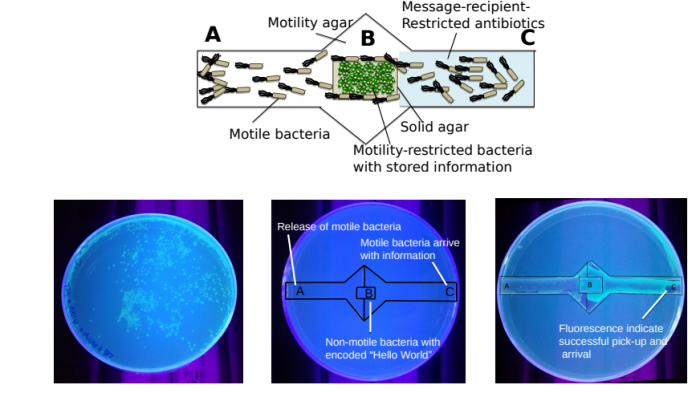
The idea is to store data in plasmids inside bacterial cells that are trapped in a specific location. To retrieve this information, the researchers send motile bacteria to this site, where they conjugate with the trapped bacteria and capture the data-carrying plasmids. Finally, the motile bacteria carry this information to a device that extracts the plasmids and reads the data they carry.
Tavella and co have even performed a proof-of-principle experiment, using two different strains of E. coli—HB101 and Novablue—that are resistant to different antibiotics. HB101 is resistant to streptomycin, while Novablue has tetracycline-resistant plasmids. Novablue can pass on this resistance to HB101 by transferring these plasmids during conjugation.
That gives the team control over where the bacteria can grow. For example, Novablue can survive when tetracycline is present, but HB101 cannot—unless it has conjugated with Novablue and become resistant.
So the prototype memory consists of a data storage area, a data reader, and a data transfer channel that connects them. To store data, the researchers encode a simple message into the tetracycline-resistant plasmids carried by the Novablue bacteria. In keeping with tradition, the message is “Hello World.” They also include a fluorescent dye in the plasmid so they can monitor its movement.
To start, the Novablue bacteria are placed in the data storage area, where they cannot escape. In practice, this is a flat surface of hard agar that is not suitable for bacterial motility. In any case, the team surrounds this with streptomycin, which kills Novablue.
The data transfer channel runs from a source of HB101 bacteria across the data storage area and then on toward the data reader. This consists of soft agar that is suitable for bacterial motility. And since HB101 is resistant to streptomycin, it can move through this channel with relative ease.
However, the region between the data storage area and the data reader is rich in tetracycline as well as streptomycin. And this prevents both bacteria from traveling across it.
What happens next is key. The HB101 bacteria travel to the data storage area, conjugate with the Novablue bacteria, and pick up the data-carrying plasmids.
But this also gives them tetracycline resistance. And that means that when they have picked up the data, they can then travel on through the channel to the data reader. The researchers then extract the plasmids and read the data—“Hello World.” They can watch the way information flows across this network thanks to the fluorescent dye.
It’s not exactly fast: the HB101 bacteria take some 72 hours to travel across the agar channel. So data rates are snail-like. But the experiment shows how a DNA data archive could work in principle.
There is another important element of a data archive. In such a system, there will be many data storage locations, and each one will have to be addressable. In other words, there must be a way for the data transfer bacteria to find each location.
Tavella and co have an answer to this too: a molecular positioning system that is analogous to the Global Positioning System. This relies on beacons that each release a chemical that attracts the bacteria. Indeed, the bacteria can be engineered to follow these chemical trails.
Then, with three different chemical trails, it is possible to triangulate a position in space. When motile bacteria follow all three trails, they end up at the location where all three chemical signals overlap. In simulations, the researchers say, this process works well, but they have yet to try it in a wet lab.
Nevertheless, the work is an interesting step towards practical DNA-based data storage. “Our solution allows digitally encoded information to be stored into non-motile bacteria, which compose an archival architecture of clusters, and to be later retrieved by engineered motile bacteria, whenever reading operations are needed,” say Tavella and co.
And the proof-of-principle experiment shows how this could work. “We have conducted wet lab experiments that show how bacteria nanonetworks can effectively retrieve a simple message, such as ‘Hello World,’ by conjugation with non-motile bacteria, and finally mobilize towards a final point,” they say.
Of course, there are many challenges ahead. The molecular positioning system is interesting but will need to be tested in a wet lab to see how versatile and practical it can be. And data rates will need to be ramped up. That won’t be possible by increasing the speed at which bacteria travel, but rates could be significantly improved by increasing the amount of data each plasmid stores.
Early days for a potentially exciting technique.
Ref: arxiv.org/abs/1801.04774 : DNA Molecular Storage System: Transferring Digitally Encoded Information through Bacterial Nanonetworks
Deep Dive
Biotechnology and health
How scientists traced a mysterious covid case back to six toilets
When wastewater surveillance turns into a hunt for a single infected individual, the ethics get tricky.
An AI-driven “factory of drugs” claims to have hit a big milestone
Insilico is part of a wave of companies betting on AI as the "next amazing revolution" in biology
The quest to legitimize longevity medicine
Longevity clinics offer a mix of services that largely cater to the wealthy. Now there’s a push to establish their work as a credible medical field.
There is a new most expensive drug in the world. Price tag: $4.25 million
But will the latest gene therapy suffer the curse of the costliest drug?
Stay connected
Get the latest updates from
MIT Technology Review
Discover special offers, top stories, upcoming events, and more.