10 Breakthrough Technologies 2006
Emerging Technologies: 2006
Each year, Technology Review selects what it believes are the 10 most important emerging technologies. The winners are chosen based on the editors’ coverage of key fields. The question that we ask is simple: is the technology likely to change the world? Some of these changes are on the largest scale possible: better biofuels, more efficient solar cells, and green concrete all aim at tackling global warming in the years ahead. Other changes will be more local and involve how we use technology: for example, 3-D screens on mobile devices, new applications for cloud computing, and social television. And new ways to implant medical electronics and develop drugs for diseases will affect us on the most intimate level of all, with the promise of making our lives healthier.
10 Breakthrough Technologies
Nanomedicine
James Baker designs nanoparticles to guide drugs directly into cancer cells, which could lead to far safer treatments.Nanomedicine
Key players
Raoul Kopelman – Nanoparticles for cancer imaging and therapy University of Michigan; Robert Langer – Nanoparticle drug delivery for prostate cancer MIT; Charles Lieber – Nanowire devices for virus detection and cancer screening Harvard University; Ralph Weissleder – Magnetic nano-particles for cancer imaging Harvard University
The treatment begins with an injection of an unremarkable-looking clear fluid. Invisible inside, however, are particles precisely engineered to slip past barriers such as blood vessel walls, latch onto cancer cells, and trick the cells into engulfing them as if they were food. These Trojan particles flag the cells with a fluorescent dye and simultaneously destroy them with a drug.
Developed by University of Michigan physician and researcher James Baker, these multipurpose nanoparticles – which should be ready for patient trials later this year – are at the leading edge of a nanotechnology-based medical revolution. Such methodically designed nanoparticles have the potential to transfigure the diagnosis and treatment of not only cancer but virtually any disease. Already, researchers are working on inexpensive tests that could distinguish a case of the sniffles from the early symptoms of a bioterror attack, as well as treatments for disorders ranging from rheumatoid arthritis to cystic fibrosis. The molecular finesse of nanotechnology, Baker says, makes it possible to “find things like tumor cells or inflammatory cells and get into them and change them directly.”
[To view an illustration of nanoparticles delivering a drug, click here.]
Cancer therapies may be the first nanomedicines to take off. Treatments that deliver drugs to the neighborhood of cancer cells in nanoscale capsules have recently become available for breast and ovarian cancers and for Kaposi’s sarcoma. The next generation of treatments, not yet approved, improves the drugs by delivering them inside individual cancer cells. This generation also boasts multifunction particles such as Baker’s; in experiments reported last June, Baker’s particles slowed and even killed human tumors grown in mice far more efficiently than conventional chemotherapy.
“The field is dramatically expanding,” says Piotr Grodzinski, program director of the National Cancer Institute’s Alliance for Nanotechnology in Cancer. “It’s not an evolutionary technology; it’s a disruptive technology that can address the problems which former approaches couldn’t.”
The heart of Baker’s approach is a highly branched molecule called a dendrimer. Each dendrimer has more than a hundred molecular “hooks” on its surface. To five or six of these, Baker connects folic-acid molecules. Because folic acid is a vitamin, most cells in the body have proteins on their surfaces that bind to it. But many cancer cells have significantly more of these receptors than normal cells. Baker links an anticancer drug to other branches of the dendrimer; when cancer cells ingest the folic acid, they consume the deadly drugs as well.
The approach is versatile. Baker has laden the dendrimers with molecules that glow under MRI scans, which can reveal the location of a cancer. And he can hook different targeting molecules and drugs to the dendrimers to treat a variety of tumors. He plans to begin human trials later this year, potentially on ovarian or head and neck cancer.
Mauro Ferrari, a professor of internal medicine, engineering, and materials science at Ohio State University, is hopeful about what Baker’s work could mean for cancer patients. “What Jim is doing is very important,” he says. “It is part of the second wave of approaches to targeted therapeutics, which I think will have tremendous acceleration of progress in the years to come.”
To hasten development of nano-based therapies, the NCI alliance has committed $144.3 million to nanotech-related projects, funding seven centers of excellence for cancer nanotechnology and 12 projects to develop diagnostics and treatments, including Baker’s.
Baker has already begun work on a modular system in which dendrimers adorned with different drugs, imaging agents, or cancer-targeting molecules could be “zipped together.” Ultimately, doctors might be able to create personalized combinations of nanomedicines by simply mixing the contents of vials of dendrimers.
Such a system is at least 10 years away from routine use, but Baker’s basic design could be approved for use in patients in as little as five years. That kind of rapid progress is a huge part of what excites doctors and researchers about nanotechnology’s medical potential. “It will completely revolutionize large branches of medicine,” says Ferrari.
Epigenetics
Alexander Olek has developed tests to detect cancer early by measuring its subtle DNA changes.Epigenetics
Key players
Stephan Beck – Epigenetics of the immune system at Wellcome Trust Sanger Institute, Cambridge, England; Joseph Bigley – Cancer diagnosis and drug development at OncoMethylome Sciences, Durham, NC; Thomas Gingeras – Gene chips for epigenetics at Affymetrix, Santa Clara, CA
Sequencing the human genome was far from the last step in explaining human genetics. Researchers still need to figure out which of the 20,000-plus human genes are active in any one cell at a given moment. Chemical modifications can interfere with the machinery of protein manufacture, shutting genes down directly or making chromosomes hard to unwind. Such chemical interactions constitute a second order of genetics known as epigenetics.
In the last five years, researchers have developed the first practical tools for identifying epigenetic interactions, and German biochemist Alexander Olek is one of the trailblazers. In 1998, Olek founded Berlin-based Epigenomics to create a rapid and sensitive test for gene methylation, a common DNA modification linked to cancer. The company’s forthcoming tests will determine not only whether a patient has a certain cancer but also, in some cases, the severity of the cancer and the likelihood that it will respond to a particular treatment. “Alex has opened up a whole new way of doing diagnostics,” says Stephan Beck, a researcher at the Wellcome Trust Sanger Institute in Cambridge, England, and an epigenetics pioneer.
Methylation adds four atoms to cytosine, one of the four DNA “letters,” or nucleotides. The body naturally uses methylation to turn genes on and off: the additional atoms block the proteins that transcribe genes. But when something goes awry, methylation can unleash a tumor by silencing a gene that normally keeps cell growth in check. Removing a gene’s natural methylation can also render a cell cancerous by activating a gene that is typically “off” in a particular tissue.
The problem is that methylated genes are hard to recognize in their native state. But Olek says Epigenomics has developed a method to detect as little as three picograms of methylated DNA; it will spot as few as three cancer cells in a tissue sample.
To create a practical diagnostic test for a given cancer, Epigenomics compares several thousand genes from cancerous and healthy cells, identifying changes in the methylation of one or more genes that correlate with the disease. Ultimately, the test examines the methylation states of only the relevant genes. The researchers go even further through a sort of epigenetic archeology: by examining the DNA in tissues from past clinical trials, they can identify the epigenetic signals in the patients who responded best or worst to a given treatment.
Philip Avner, an epigenetics pioneer at the Pasteur Institute in Paris, says that Epigenomics’ test is a powerful tool for accurately diagnosing and understanding cancers at their earliest stages. “If we can’t prevent cancer, at least we can treat it better,” says Avner.
Roche Diagnostics expects to bring Epigenomics’ first product, a screening test for colon cancer, to market in 2008. The test is several times more likely to spot a tumor than the current test, which measures the amount of blood in a stool sample. And thanks to the sensitivity of its process, Epigenomics can detect the tiny amounts of methylated DNA such tumors shed into the bloodstream, so only a standard blood sample is required. The company is working on diagnostics for three more cancers: non-Hodgkin’s lymphoma, breast cancer, and prostate cancer.
Olek believes that epigenetics could also have applications in helping explain how lifestyle affects the aging process. It might reveal, for example, why some individuals have a propensity toward diabetes or heart disease.
Olek’s goal is a human-epigenome mapping project that would identify the full range of epigenetic variation possible in the human genome. Such a map, Olek believes, could reveal the missing links between genetics, disease, and the environment. Today, progress on the methylation catalogue is accelerating, thanks to Epigenomics and the Wellcome Trust Sanger Institute, which predict that the methylation status of 10 percent of human genes will be mapped by the end of this year.
Cognitive Radio
To avoid future wireless traffic jams, Heather “Haitao” Zheng is finding ways to exploit unused radio spectrum.Cognitive Radio
Key players
Bob Broderson – Advanced communication algorithms and low-power devices at University of California, Berkeley; John Chapin – Software-defined radios at Vanu, Cambridge, MA; Michael Honig – Pricing algorithm for spectrum sharing at Northwestern University; Joseph Mitola III – Cognitive radios at Mitre, McLean, VA; Adam Wolisz – Protocols for communications networks at Technical University of Berlin, Germany
Growing numbers of people are making a habit of toting their laptops into Starbuck’s, ordering half-caf skim lattes, and plunking down in chairs to surf the Web wirelessly. That means more people are also getting used to being kicked off the Net as computers competing for bandwidth interfere with one another. It’s a local effect – within 30 to 60 meters of a transceiver – but there’s just no more space in the part of the radio spectrum designated for Wi-Fi.
Imagine, then, what happens as more devices go wireless – not just laptops, or cell phones and BlackBerrys, but sensor networks that monitor everything from temperature in office buildings to moisture in cornfields, radio frequency ID tags that track merchandise at the local Wal-Mart, devices that monitor nursing-home patients. All these gadgets have to share a finite – and increasingly crowded – amount of radio spectrum.
Heather Zheng, an assistant professor of computer science at the University of California, Santa Barbara, is working on ways to allow wireless devices to more efficiently share the airwaves. The problem, she says, is not a dearth of radio spectrum; it’s the way that spectrum is used.
The Federal Communications Commission in the United States, and its counterparts around the world, allocate the radio spectrum in swaths of frequency of varying widths. One band covers AM radio, another VHF television, still others cell phones, citizen’s-band radio, pagers, and so on; now, just as wireless devices have begun proliferating, there’s little left over to dole out.
But as anyone who has twirled a radio dial knows, not every channel in every band is always in use. In fact, the FCC has determined that, in some locations or at some times of day, 70 percent of the allocated spectrum may be sitting idle, even though it’s officially spoken for.
Zheng thinks the solution lies with cognitive radios, devices that figure out which frequencies are quiet and pick one or more over which to transmit and receive data. Without careful planning, however, certain bands could still end up jammed. Zheng’s answer is to teach cognitive radios to negotiate with other devices in their vicinity. In Zheng’s scheme, the FCC-designated owner of the spectrum gets priority, but other devices can divvy up unused spectrum among themselves.
But negotiation between devices uses bandwidth in itself, so Zheng simplified the process. She selected a set of rules based on “game theory” – a type of mathematical modeling often used to find the optimal solutions to economics problems – and designed software that made the devices follow those rules. Instead of each radio’s having to tell its neighbor what it’s doing, it simply observes its neighbors to see if they are transmitting and makes its own decisions.
Zheng compares the scheme to a driver’s reacting to what she sees other drivers doing. “If I’m in a traffic lane that is heavy, maybe it’s time for me to shift to another lane that is not so busy,” she says. When shifting lanes, however, a driver needs to follow rules that prevent her from bumping into others.
Zheng has demonstrated her approach in computer simulations and is working toward testing it on actual hardware. But putting spectrum-sharing theory into practice will take engineering work, from designing the right antennas to writing the software that will run the cognitive radios, Zheng acknowledges. “This is just a starting phase,” she says.
Nonetheless, cognitive radios are already making headway in the real world. Intel has plans to build reconfigurable chips that will use software to analyze their environments and select the best protocols and frequencies for data transmission. The FCC has made special allowances so that new types of wireless networks can test these ideas on unused television channels, and the Institute of Electrical and Electronics Engineers, which sets many of the technical standards that continue to drive the Internet revolution, has begun considering cognitive-radio standards.
It may be 10 years before all the issues get sorted out, Zheng says, but as the airwaves become more crowded, all wireless devices will need more-efficient ways to share the spectrum.
Nuclear Reprogramming
Hoping to resolve the embryonic-stem-cell debate, Markus Grompe envisions a more ethical way to derive the cells.Embryonic stem cells may spark more vitriolic argument than any other topic in modern science. Conservative Christians aver that the cells’ genesis, which requires destroying embryos, should make any research using them taboo. Many biologists believe that the cells will help unlock the secrets of devastating diseases such as Parkinson’s and multiple sclerosis, providing benefits that far outweigh any perceived ethical harm.
 BRYAN CHRISTIE DESIGN
BRYAN CHRISTIE DESIGNNuclear Reprogramming
Key players
George Daley – Studying nanog’s ability to reprogram nuclei at Harvard Medical Schoo; Kevin Eggan – Reprogramming adult cells using stem cells at Harvard University; Rudolf Jaenisch – Creating tailored stem cells using altered nuclear transfer (CDX2) at MIT
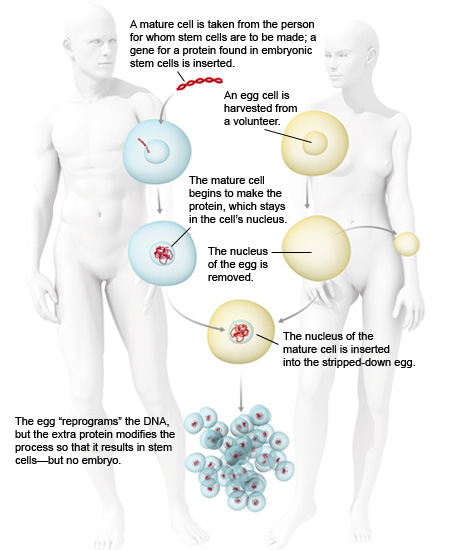
Markus Grompe, director of the Oregon Stem Cell Center at Oregon Health and Science University in Portland, hopes to find a way around the debate by producing cloned cells that have all the properties of embryonic stem cells – but don’t come from embryos.
His plan involves a variation on the cloning procedure that produced Dolly the sheep. In the original procedure, scientists transferred the genetic material from an adult cell into an egg stripped of its own DNA. The egg’s proteins reprogrammed the adult DNA, creating an embryo genetically identical to the adult donor. Grompe believes that by forcing the donor cell to produce a protein called nanog, which is normally found only in embryonic stem cells, he can alter the reprogramming process so that it never results in an embryo. Instead, it would yield a cell with many of the characteristics of an embryonic stem cell.
Grompe’s work is part of a growing effort to find alternative ways to create cells with the versatility of embryonic stem cells. Many scientists hope to use proteins to directly reprogram, say, skin cells to behave like stem cells.
Others think smaller molecules may do the trick; Scripps Research Institute chemist Peter Schultz has found a chemical that turns mouse muscle cells into cells able to form fat and bone cells. And Harvard University biologist Kevin Eggan believes it may be possible to create stem cells whose DNA matches a specific patient’s by using existing stem cells stripped of their DNA to reprogram adult cells.
Meanwhile, researchers have tested methods for extracting stem cells without destroying viable embryos. Last fall, MIT biologist Rudolf Jaenisch and graduate student Alexander Meissner showed that by turning off a gene called CDX2 in the nucleus of an adult cell before transferring it into a nucleus-free egg cell, they could create a biological entity unable to develop into an embryo – but from which they could still derive normal embryonic stem cells.
Also last fall, researchers at Advanced Cell Technology in Worcester, MA, grew embryonic stem cells using a technique that resembles something called preimplantation genetic diagnosis (PGD). PGD is used to detect genetic abnormalities in embryos created through in vitro fertilization; doctors remove a single cell from an eight-cell embryo for testing. The researchers separated single cells from eight-cell mouse embryos, but instead of testing them, they put each in a separate petri dish, along with embryonic stem cells. Unidentified factors caused the single cells to divide and develop some of the characteristics of stem cells. When the remaining seven-cell embryos were implanted into female mice, they developed into normal mice.
Such methods, however, are unlikely to resolve the ethical debate because, in the eyes of some, they still endanger embryos. Grompe’s approach holds out the promise of unraveling the moral dilemma. If it works, no embryo will have been produced – so no potential life will be harmed. As a result, some conservative ethicists have endorsed Grompe’s proposal.
Whether it is actually a feasible way to harvest embryonic stem cells remains uncertain. Some are skeptical. “There’s really no evidence it would work,” says Jaenisch. “I doubt it would.” But the experiments Grompe proposes, Jaenisch says, would still be scientifically valuable in helping explain how to reprogram cells to create stem cells. Harvard Stem Cell Institute scientist George Daley agrees. In fact, Daley’s lab is also studying nanog’s ability to reprogram adult cells.
Still, many biologists and bioethicists have mixed feelings about efforts to reprogram adult cells to become pluripotent. While they agree the research is important, they worry that framing it as a search for a stem cell compromise may slow funding – private and public – for embryonic-stem-cell research, hampering efforts to decipher or even cure diseases that affect thousands of desperate people. Such delays, they argue, are a greater moral wrong than the loss of cells that hold only the potential for life.
Many ethicists – and the majority of Americans – seem to agree. “We’ve already decided as a society that it’s perfectly okay to create and destroy embryos to help infertile couples to have babies. It seems incredible to me that we could say that that’s a legitimate thing to do, but we can’t do the same thing to help fight diseases that kill children,” says David Magnus, director of the Stanford Center for Biomedical Ethics.
Diffusion Tensor Imaging
Kelvin Lim is using a new brain-imaging method to understand schizophrenia.Flipping through a pile of brain scans, a neurologist or psychiatrist would be hard pressed to pick out the one that belonged to a schizophrenic. Although schizophrenics suffer from profound mental problems – hallucinated conversations and imagined conspiracies are the best known – their brains look more or less normal.
This contradiction fascinated Kelvin Lim, a neuroscientist and psychiatrist at the University of Minnesota Medical School, when he began using imaging techniques such as magnetic resonance imaging (MRI) to study the schizophrenic brain in the early 1990s. Lim found subtle hints of brain structures gone awry, but to understand how these problems led to the strange symptoms of schizophrenia, he needed a closer look at the patients’ neuroanatomy than standard scans could provide.
Then, in 1996, a colleague told him about diffusion tensor imaging (DTI), a newly developed variation of MRI that allowed scientists to study the connections between different brain areas for the first time.
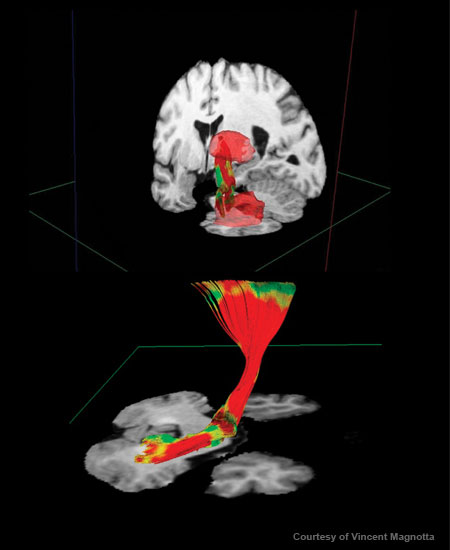
Diffusion tensor imaging (DTI) yields images of nerve fiber tracts; different colors indicate the organization of the nerve fibers. Here, a tract originating at the cerebellum is superimposed on a structural-MRI image of a cross section of the brain. Diffusion Tensor Imaging
Key players
Peter Basser – Development of higher-resolution diffusion imaging techniques at National Institute of Child Health and Human Development; Aaron Field – Neurosurgery planning at University of Wisconsin-Madison; Michael Moseley – Assessment and early treatment of stroke at Stanford University
Lim has pioneered the use of DTI to understand psychiatric disease. He was one of the first to use the technology to uncover minute structural aberrations in the brains of schizophrenics. His group has recently found that memory and cognitive problems associated with schizophrenia, major but undertreated aspects of the disease, are linked to flaws in nerve fibers near the hippocampus, a brain area crucial for learning and memory. “DTI allows us to examine the brain in ways we hadn’t been able to before,” says Lim.
Conventional imaging techniques, such as structural MRI, reveal major anatomical features of the brain – gray matter, which is made up of nerve cell bodies. But neuroscientists believe that some diseases may be rooted in subtle “wiring” problems involving axons, the long, thin tails of neurons that carry electrical signals and constitute the brain’s white matter. With DTI, researchers can, for the first time, look at the complex network of nerve fibers connecting the different brain areas. Lim and his colleagues hope this sharper view of the brain will help better define neurological and psychiatric diseases and yield more-targeted treatments.
In DTI, radiologists use specific radio-frequency and magnetic field-gradient pulses to track the movement of water molecules in the brain. In most brain tissue, water molecules diffuse in all different directions. But they tend to diffuse along the length of axons, whose coating of white, fatty myelin holds them in. Scientists can create pictures of axons by analyzing the direction of water diffusion.
Following Lim’s lead, other neuroscientists have begun using DTI to study a host of disorders, including addiction, epilepsy, traumatic brain injury, and various neurodegenerative diseases. For instance, DTI studies have shown that chronic alcoholism degrades the white-matter connections in the brain, which may explain the cognitive problems seen in heavy drinkers. Other DTI projects are examining how the neurological scars left by stroke, multiple sclerosis, and amyotrophic lateral sclerosis (better known as Lou Gehrig’s disease) are linked to patients’ disabilities.
Lim is pushing the technology even further by combining it with findings from other fields, such as genetics, to unravel the mysteries of neurological and psychiatric disorders. Lim’s group has found, for instance, that healthy people with a genetic risk for developing Alzheimer’s disease have tiny structural defects in specific parts of the brain that are not shared by noncarriers. How these defects might be linked to the neurological problems of Alzheimer’s isn’t clear, but the researchers are trying to find the connection.
Lim and others also continue to refine DTI itself, striving for an even closer look at the brain’s microarchitecture. For example, current DTI techniques can easily image brain areas with large bundles of fibers all moving in the same direction, such as the corpus callosum, which connects the two hemispheres of the brain. But it has difficulty with areas such as the one where fibers leave the corpus callosum for other parts of the brain, which is a tangle of wires.
Researchers hope tools for studying white matter, like DTI, will help illuminate the mysteries of both healthy and diseased brains. Lim believes his own research into diseases like schizophrenia and Alzheimer’s could yield better diagnostics within 10 to 20 years – providing new hope for the next generation of patients.
Universal Authentication
Leading the development of a privacy-protecting online ID system, Scott Cantor is hoping for a safer Internet.If you’re like most people, you’ve established multiple user IDs and passwords on the Internet – for your employer or school, your e-mail accounts, online retailers, banks, and so forth. It’s cumbersome and confusing, slowing down online interactions if only because it’s so easy to forget the plethora of passwords. Worse, the diversity of authentication systems increases the chances that somewhere, your privacy will be compromised, or your identity will be stolen.
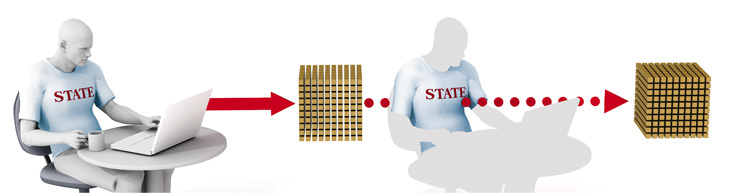
Security with privacy: Shibboleth software could create a far more trustworthy Internet by allowing a one-step login that carries through to many online organizations, confirming identity but preserving privacy. In this example, a student logs in to his university's site, then clicks through to a second university. Shibboleth confirms that the person is a student but doesn't give his name. BRYAN CHRISTIE DESIGNUniversal Authentication
Key players
Stefan Brands – Cryptology, identity management, and authentication technologies at McGill University; Kim Cameron – “InfoCard” system to manage and employ a range of digital identity information at Microsoft, Redmond, WA; Robert Morgan – “Person registry” that gathers identity data from source systems; scalable authentication infrastructure at University of Washington; Tony Nadalin – Personal-identity software platform at IBM, Armonk, NY
The balkanization of today’s online identity-verifying systems is a big part of the Internet’s fraud and security crisis. As Kim Cameron, Microsoft’s architect of identity and access, puts it in his blog, “If we do nothing, we will face rapidly proliferating episodes of theft and deception that will cumulatively erode public trust in the Internet.” Finding ways to bolster that trust is critically important to preserving the Internet as a useful, thriving medium, argues David D. Clark, an MIT computer scientist and the Internet’s onetime chief protocol architect.
Scott Cantor, a senior systems developer at Ohio State University, thinks the answer may lie in Web “authentication systems” that allow users to hop securely from one site to another after signing on just once. Such systems could protect both users’ privacy and the online businesses and other institutions that offer Web-based services.
Cantor led the technical development of Shibboleth, an open-standard authentication system used by universities and the research community, and his current project is to expand its reach. He has worked, not only to make the system function smoothly, but also to build bridges between it and parallel corporate efforts. “Scott is the rock star of the group,” says Steven Carmody, an IT architect at Brown University who manages a Shibboleth project for Internet2, an Ann Arbor, MI-based research consortium that develops advanced Internet technologies for research laboratories and universities. “Scott’s work has greatly simplified the management of these Internet-based relationships, while ensuring the required security and level of assurance for each transaction.”
Shibboleth acts not only as an authentication system but also – counterintuitively – as a guardian of privacy. Say a student at Ohio State wishes to access Brown’s online library. Ohio State securely holds her identifying information – name, age, campus affiliations, and so forth. She enters her user ID and password into a page on Ohio State’s website. But when she clicks through to Brown, Shibboleth takes over. It delivers only the identifying information Brown really needs to know: the user is a registered Ohio State student.
While some U.S. universities have been using Shibboleth since 2003, adoption of the system grew rapidly in 2005. It’s now used at 500-plus sites worldwide, including educational systems in Australia, Belgium, England, Finland, Denmark, Germany, Switzerland, and the Netherlands; even institutions in China are signing on. Also in late 2005, Internet2 announced Shibboleth’s interoperability with a Microsoft security infrastructure called the Active Directory Federation Service.
Critically, the system is moving into the private sector, too. The science and medical division of research publishing conglomerate Reed Elsevier has begun granting university-based subscribers access to its online resources through Shibboleth, rather than requiring separate, Elsevier-specific logins. And Cantor has forged ties with the Liberty Alliance, a consortium of more than 150 companies and other institutions dedicated to creating shared identity and authentication systems.
With Cantor’s help, the alliance, which includes companies such as AOL, Bank of America, IBM, and Fidelity Investments, is basing the design of its authentication systems on a common standard known as SAML. The alliance, Cantor says, was “wrestling with lots of the same hard questions that we were, and we were starting to play in the same kind of territories. Now there is a common foundation….we’re trying to make it ubiquitous.” With technical barriers overcome, the companies can now roll out systems as their business needs dictate.
Of course, Cantor is not the only researcher, nor Shibboleth the only technology, in the field of Internet authentication. In 1999, for instance, Microsoft launched its Passport system, which let Windows users access any participating website using their e-mail addresses and passwords. Passport, however, encountered a range of security and privacy problems.
But thanks to the efforts of the Shibboleth team and the Liberty Alliance, Web surfers could start accessing multiple sites with a single login in the next year or so, as companies begin rolling out interoperable authentication systems.
Nanobiomechanics
Measuring the tiny forces acting on cells, Subra Suresh believes, could produce fresh understanding of diseases.Most people don’t think of the human body as a machine, but Subra Suresh does. A materials scientist at MIT, Suresh measures the minute mechanical forces acting on our cells.
Medical researchers have long known that diseases can cause – or be caused by – physical changes in individual cells. For instance, invading parasites can distort or degrade blood cells, and heart failure can occur as muscle cells lose their ability to contract in the wake of a heart attack. Knowing the effect of forces as small as a piconewton – a trillionth of a newton – on a cell gives researchers a much finer view of the ways in which diseased cells differ from healthy ones.
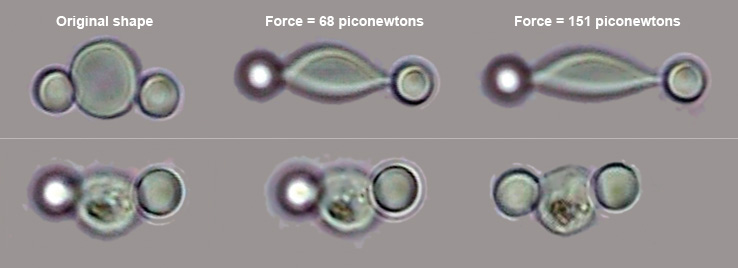
Optical tweezers stretch a healthy red blood cell (top row), increasing the applied force slowly, by a matter of piconewtons. A cell in a late stage of malarial infection is stretched in a similar fashion (bottom row). The experiment illustrates how the infected cell becomes rigid, which prevents it from traveling easily through blood capillaries and helps cause the symptoms of malaria. SUBRA SURESHNanobiomechanics
Key players
Eduard Arzt – Structure and mobility of pancreatic cancer cells Max Planck at Institute, Stuttgart, Germany; Peter David and Genevieve Milon – Parasite-host interaction; mechanics of the spleen at Pasteur Institute, Paris, France; Ju Li – Models of internal cellular structures at Ohio State University; C. T. Lim and Kevin Tan – Red-blood-cell mechanics at National University of Singapore
Suresh spent much of his career making nanoscale measurements of materials such as the thin films used in microelectronic components. But since 2003, Suresh’s laboratory has spent more and more time applying nanomeasurement techniques to living cells. He’s now among a pioneering group of materials scientists who work closely with microbiologists and medical researchers to learn more about how our cells react to tiny forces and how their physical form is affected by disease. “We bring to the table expertise in measuring the strength of materials at the smallest of scales,” says Suresh.
One of Suresh’s recent studies measured mechanical differences between healthy red blood cells and cells infected with malaria parasites. Suresh and his collaborators knew that infected blood cells become more rigid, losing the ability to reduce their width from eight micrometers down to two or three micrometers, which they need to do to slip through capillaries. Rigid cells, on the other hand, can clog capillaries and cause cerebral hemorrhages. Though others had tried to determine exactly how rigid malarial cells become, Suresh’s instruments were able to bring greater accuracy to the measurements. Using optical tweezers, which employ intensely focused laser light to exert a tiny force on objects attached to cells, Suresh and his collaborators showed that red blood cells infected with malaria become 10 times stiffer than healthy cells – three to four times stiffer than was previously estimated.
Eduard Arzt, director of materials research at the Max Planck Institute in Stuttgart, Germany, says that Suresh’s work is important because cell flexibility is a vital characteristic not only of malarial cells but also of metastasizing cancer cells. “Many of the mechanical concepts we’ve been using for a long time, like strength and elasticity, are also very important in biology,” says Arzt.
Arzt and Suresh both caution that it’s too early to say that understanding the mechanics of human cells will lead to more effective treatments. But what excites them and others in the field is the ability to measure the properties of cells with unprecedented precision. That excitement seems to be spreading: in October, Suresh helped inaugurate the Global Enterprise for Micro-Mechanics and Molecular Medicine, an international consortium that will use nanomeasurement tools to tackle major health problems, including malaria, sickle-cell anemia, cancer of the liver and pancreas, and cardiovascular disease. Suresh serves as the organization’s founding director.
“We know mechanics plays a role in disease,” says Suresh. “We hope it can be used to figure out treatments.” If it can, the tiny field of nanomeasurement could have a huge impact on the future of medicine.
Pervasive Wireless
Can’t all our wireless gadgets just get along? It’s a question that Dipankar Raychaudhuri is trying to answer.Pervasive Wireless
Key players
David Culler – Operating systems and middleware for wireless sensors at University of California, Berkeley; Kazuo Imai – Integrating cellular with other network technology at NTT DoCoMo, Tokyo, Japan; Lakshman Krishnamurthy and Steven Conner – Wireless network architecture at Intel, Santa Clara, CA
In New Brunswick, NJ, is a large, white room with an army of yellow boxes hanging from the ceiling. Eight hundred in all, the boxes are actually a unique grid of radios that lets researchers design and test ways to link mobile, radio-equipped computers in configurations that can change on the fly.
The ability to form such ad hoc networks, says Dipankar Raychaudhuri, director of the Rutgers University lab that houses the radios, will be critical to the advent of pervasive computing–in which everything from your car to your coffee cup “talks” to other devices in an attempt to make your life run more smoothly.
Wireless transactions already take place; anybody who speeds through tolls with an E-ZPass transmitter participates in them daily. But Raychaudhuri foresees a not-too-distant day when radio frequency identification (RFID) tags embedded in merchandise call your cell phone to alert you to sales, cars talk to each other to avoid collisions, and elderly people carry heart and blood-pressure monitors that can call a doctor during a medical emergency. Even mesh networks, collections of wireless devices that pass data one to another until it reaches a central computer, may need to be connected to pagers, cell phones, or other gadgets that employ diverse wireless protocols.
Hundreds of researchers at universities, large companies such as Microsoft, Intel, and Nortel, and small startups are developing embedded radio devices and sensors. But making computing truly pervasive entails tying these disparate pieces together, says Raychaudhuri, a professor of electrical and computer engineering at Rutgers. Finding ways to do that is what the radio test grid, which Raychaudhuri built with computer scientists Ivan Seskar and Max Ott, is for.
One problem the researchers are addressing is that different devices communicate using different radio standards: RFID tags use one set of standards, cell phones still others, and various Wi-Fi devices several versions of a third. Linking such devices into a pervasive network means providing them with a common protocol.
Take, for example, the issue of automotive safety. Enabling cars to communicate with each other could prevent crashes; in Raychaudhuri’s vision, each car would have a Global Positioning System unit and send its exact location to nearby vehicles. But realizing that vision requires a protocol that allows the cars not only to communicate but also to decide how many other cars they should include in their networks and how close another car should be to be included. As programmers develop candidates for such a protocol, they try them out on the radio test bed. Each yellow box contains a computer and three different radios, two for handling the various Wi-Fi standards and one that uses either Bluetooth or ZigBee, short-range wireless protocols for personal electronics and for monitoring or control devices, respectively. The researchers configure the radios to mimic the situation they want to test and load their protocols to see, for instance, how long it takes each radio to detect neighbors and send data. “If I want cars not to collide, it cannot take 10 seconds to determine that a car is nearby,” says Raychaudhuri. “It has to take a few microseconds.”
The Rutgers radio grid is the first large-scale shared research facility that researchers can use to study multiple wireless devices and network technologies. “The sort of real-world complexity, dealing with real-world numbers that [the test bed] allows you to do, is something that really makes it quite unique,” says Tod Sizer, director of the Wireless Technology Research Department at Lucent Technologies’ Bell Labs.
Sizer’s group is working with Raychaudhuri to build cognitive-radio boxes that can be programmed to employ a wide variety of wireless standards, such as RFID, Wi-Fi, or cellular-phone protocols.
While hordes of researchers are developing new networked devices, Raychaudhuri says it is the standardization of communications protocols that will make pervasive computing take off. In just five years, he believes, networks of embedded devices will be all around us. His aim is to reduce “friction” in daily life, eliminating lines, saving time in searching for objects, automating security checkpoints in airports, and the like. “You save 10 seconds here, two minutes there, but it’s significant,” he says. He claims that just a 2 percent reduction of friction in the world’s economy could be worth hundreds of billions of dollars in productivity. “Each transaction is small, but the benefit to society is very large.”
Stretchable Silicon
By teaching silicon new tricks, John Rogers is reinventing the way we use electronics.These days, most electronic circuitry comes in the form of rigid chips, but devices thin and flexible enough to be rolled up like a newspaper are fast approaching. Already, “smart” credit cards carry bendable microchips, and companies such as Fujitsu, Lucent Technologies, and E Ink are developing “electronic paper” – thin, paperlike displays.
But most truly flexible circuits are made of organic semiconductors sprayed or stamped onto plastic sheets. Although useful for roll-up displays, organic semiconductors are just too slow for more intense computing tasks. For those jobs, you still need silicon or another high-speed inorganic semiconductor. So John Rogers, a materials scientist at the University of Illinois at Urbana-Champaign, found a way to stretch silicon.
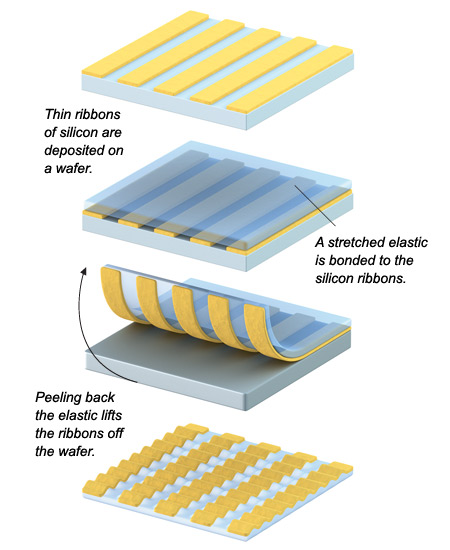
Releasing the tension on the elastic produces “waves” of silicon that can later be stretched out again as needed. Such flexible silicon could be used to make wearable electronics. BRYAN CHRISTIE DESIGNStretchable Silicon
Key players
Stephanie Lacour – Neuro-electronic prosthesis to repair damage to the nervous system at University of Cambridge, England; Takao Someya – Large-area electronics based on organic transistors at University of Tokyo; Sigurd Wagner – Electronic skin based on thin-film silicon at Princeton University
If bendable is good, stretchable is even better, says Rogers, especially for high-performance conformable circuits of the sort needed for so-called smart clothes or body armor. “You don’t comfortably wear a sheet of plastic,” he says. The potential applications of circuitry made from Roger’s stretchable silicon are vast. It could be used in surgeons’ gloves to create sensors that would read chemical levels in the blood and alert a surgeon to a problem, without impairing the sense of touch. It could allow a prosthetic limb to use pressure or temperature cues to change its shape.
What makes Rogers’s work particularly impressive is that he works with single-crystal silicon, the same type of silicon found in microprocessors. Like any other single crystal, single-crystal silicon doesn’t naturally stretch. Indeed, in order for it even to bend, it must be prepared as an ultrathin layer only a few hundred nanometers thick on a bendable surface. Rogers exploits the flexibility of thin silicon, but instead of attaching it to plastic, he affixes it in narrow strips to a stretched-out, rubber-like polymer. When the stretched polymer snaps back, the silicon strips buckle but do not break, forming “waves” that are ready to stretch out again.
Rogers’s team has fabricated diodes and transistors – the basic building blocks of electronic devices – on the thin ribbons of silicon before bonding them to the polymer; the wavy devices work just as well as conventional rigid versions, Rogers says. In theory, that means complete circuits of the sort found in computers and other electronics would also work properly when rippled.
Rogers isn’t the first researcher to build stretchable electronics. A couple of years ago, Princeton University’s Sigurd Wagner and colleagues began making stretchable circuits after inventing elastic-metal interconnects. Using the stretchable metal, Wagner’s group connected together rigid “islands” of silicon transistors. Although the silicon itself couldn’t stretch, the entire circuit could. But, Wagner notes, his technique isn’t suited to making electrically demanding circuits such as those in a Pentium chip. “The big thing that John has done is use standard, high-performance silicon,” says Wagner.
Going from simple diodes to the integrated circuits needed to make sensors and other useful microchips could take at least five years, says Rogers. In the meantime, his group is working to make silicon even more flexible. When the silicon is affixed to the rubbery surface in rows, it can stretch only in one direction. By changing the strips’ geometry, Rogers hopes to make devices pliable enough to be folded up like a T-shirt. That kind of resilience could make silicon’s future in electronics stretch out a whole lot further.
Comparative Interactomics
By creating maps of the body’s complex molecular interactions, Trey Ideker is providing new ways to find drugs.Comparative Interactomics
Key players
James Collins – Synthetic gene networks at Boston University; Bernhard Palsson – Metabolic networks at University of California, San Diego; Marc Vidal – Comparison of interactomes among species at Dana-Farber Cancer Institute, Boston, MA
Biomedical research these days seems to be all about the “omes”: genomes, proteomes, metabolomes. Beyond all these lies the mother of all omes – or maybe just the ome du jour: the interactome. Every cell hosts a vast array of interactions among genes, RNA, metabolites, and proteins. The impossibly complex map of all these interactions is, in the language of systems biology, the interactome.
Trey Ideker, a molecular biotechnologist by way of electrical engineering, has recently begun comparing what he calls the “circuitry” of the interactomes of different species. “It’s really an incremental step in terms of the concepts, but it’s a major leap forward in that we can gather and analyze completely new types of information to characterize biological systems,” says Ideker, who runs the Laboratory for Integrative Network Biology at the University of California, San Diego. “I think it’s going to be cool to map out the circuitry of all these cells.”
Beyond the cool factor, Ideker and other leaders in the nascent field of interactomics hope that their work may help uncover new drugs, improve existing drugs by providing a better understanding of how they work, and even lead to computerized models of toxicity that could replace studies now conducted on animals. “Disease and drugs are about pathways,” Ideker says.
Ideker made a big splash in the field in 2001 while still a graduate student with Leroy Hood at the Institute for Systems Biology in Seattle. In a paper for Science, Ideker, Hood, and coworkers described in startling detail how yeast cells use sugar. They presented a wiring-like diagram illustrating everything from the suite of genes involved, to the protein-protein interactions, to how perturbing the system altered different biochemical pathways. “His contribution was really special,” says geneticist Marc Vidal of the Dana-Farber Cancer Institute in Boston, who introduced the concept that interactomes can be conserved between species. “He came up with one of the first good visualization tools.”
Last November, Ideker’s team turned heads by reporting in Nature that it had aggregated in one database all the available protein-protein interactomes of yeast, the fruit fly, the nematode worm, and the malaria-causing parasite Plasmodium falciparum. Though there’s nothing particularly novel about comparing proteins across species, Ideker’s lab is one of the few that has begun hunting for similarities and differences between the protein-protein interactions of widely different creatures. It turns out that the interactomes of yeast, fly, and worm include interactions called protein complexes that have some similarities between them. This conservation across species indicates that the interactions may serve some vital purpose. But Plasmodium, oddly, shares no protein complexes with worm or fly and only three with yeast. “For a while, we struggled to figure out what was going wrong with our analysis,” says Ideker. After rechecking their data, Ideker and his team concluded that Plasmodium probably just had a somewhat different interactome.
For pharmaceutical makers, the discovery of unique biological pathways, such as those found in the malaria parasite, suggests new drug targets. Theoretically, a drug that can interrupt such a pathway will have limited, if any, impact on circuits in human cells, reducing the likelihood of toxic side effects. Theoretically. In reality, pharmaceutical companies aren’t exactly tripping over themselves to make new drugs for malaria – a disease that strikes mainly in poor countries. But the general idea has great promise, says Ideker, who now plans to compare the interactomes of different HIV strains to see whether any chinks in that virus’s armor come to light.
George Church, who directs the Lipper Center for Computational Genetics at Harvard Medical School, has high respect for Ideker but adds another caveat: existing interactome data comes from fast, automated tests that simply aren’t that accurate yet. “The way I divide the omes is by asking, Are these data permanent, or are they going to be replaced by something better?” says Church. Data on the DNA sequences of genomes, Church says, is permanent. But interactome data? “There’s a 50-50 chance that this will be true or accepted in two years,” says Church. “That’s not Trey’s fault. He’s one of the people who is trying to make it more rigorous.”
Ideker agrees that “there’s a lot of noise in the system,” but he says the continuing flood of interactome data is making what happens inside different cells ever more clear. “Within five years, we hope to take these interaction data and build models of cellular circuitry to predict actions of drugs before they’re in human trials. That’s the billion-dollar application.”