The Mystery of Knotted Proteins
Proteins are long chains of amino acids that are essential building blocks for all living things. These chains form complex three-dimensional shapes that play a key part in their function—when molecules fit together like a lock and key, for example.
So one of the great challenges in molecular biology is to understand how proteins form these shapes, and how they do it so reliably and rapidly. This is the problem of protein folding.
There is an interesting subplot in this mystery. For many years, molecular biologists argued that although proteins can be highly tangled, they could not form knots under ordinary circumstances because this would trap the structure and prevent it from folding further.
Since the turn of the century, however, a different view has emerged. Biologists have discovered that some proteins do form knots. And that raises a couple of interesting questions: how do these knots form and why?
Today, we get an insight thanks to the work of Sophie Jackson at the University of Cambridge and a couple of pals. These guys review the field of knot-forming proteins and set out the major questions that remain unanswered.
This work has significant potential. Proteins that are unfolded or misfolded can have toxic effects, so a better understanding of knots and why they form could have important medical implications.
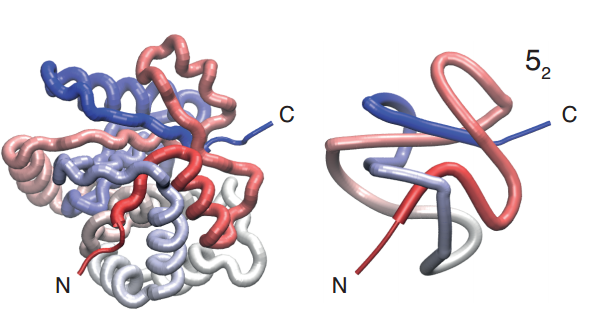
Knots are usually catalogued in terms of the number of crossings and the number of variations these crossings allow. A simple trefoil knot has three crossings with just one variation, so this is designated 31. The more complex knot with five crossings has two versions designated 51 and 52, while knots with seven crossings come in seven varieties designated 71, 72, … 77. And so on. The number of variations increases exponentially with the crossing number.
Biologists have discovered an increasing variety of knotted proteins. Indeed, about 1 percent of the entries in the Protein Data Bank are knotted, and at least 19 proteins form simple 31 trefoils.
Some of these knotted proteins play an important role in human biochemistry. For example, human ubiquitin C-terminal hydrolase isoform 1 (UCH-L1) is 52 knotted and accounts for up to 5 percent of soluble proteins in neurons.
UCH-L1 has been the focus of considerable study, not least because an unknotted version of this molecule is implicated in Parkinson’s disease. In one study, researchers used optical tweezers to create different versions of this molecule that were either unknotted, 31 knotted, or 52 knotted. They then measured how the protein refolded.
As it turns out, the presence of a knot significantly slows the rate at which a protein folds. It also creates a more complex energy landscape that allows a much wider range of intermediate shapes to form during the folding process. What’s more, the 52 knotted region ends up being much larger than it need be.
Exactly what role the additional shapes might play or why a slower folding speed may be important isn’t clear. All this will need to be explored computationally in the future. The complexity of these problems makes this hard, even for today’s most powerful computers, so better folding simulations will be an important area for future work.
One interesting clue is that knots often occur in proteins near the sites where enzymes bind to the molecule. That suggests that the knot shape forms a crucial part of the lock and key shape. This may explain their presence—knots may allow a protein to form shapes that are otherwise difficult or impossible to achieve.
Jackson and co finish by listing a set of unsolved questions in this field. Some of these are related to limitations in the way biologists can simulate knotting—for example, do these simulations miss any important steps in the knotting process?
Another challenge is to understand whether more complex structures can form out of composite knots, when one knot forms inside another. Some theoretical work suggests that these kinds of structures could have important benefits.
And finally, is it possible to change a knotted protein into an unknotted one with a few judicious cuts in the structure? That’s something that an enterprising enzyme might easily achieve.
A better understanding of the role knots play in protein folding will have important implications for biochemistry. And this kind of knowledge can also be put to good use in discovering and developing therapeutic drugs. That’s why these questions have more than academic interest.
Ref: arxiv.org/abs/1610.05779 : How to Fold Intricately: Using Theory And Experiments to Unravel the Properties of Knotted Proteins
Keep Reading
Most Popular
Large language models can do jaw-dropping things. But nobody knows exactly why.
And that's a problem. Figuring it out is one of the biggest scientific puzzles of our time and a crucial step towards controlling more powerful future models.
The problem with plug-in hybrids? Their drivers.
Plug-in hybrids are often sold as a transition to EVs, but new data from Europe shows we’re still underestimating the emissions they produce.
How scientists traced a mysterious covid case back to six toilets
When wastewater surveillance turns into a hunt for a single infected individual, the ethics get tricky.
Google DeepMind’s new generative model makes Super Mario–like games from scratch
Genie learns how to control games by watching hours and hours of video. It could help train next-gen robots too.
Stay connected
Get the latest updates from
MIT Technology Review
Discover special offers, top stories, upcoming events, and more.